IDDomainSpotter: Compositional bias reveals domains in long disordered protein regions - Insights from transcription factors
Millard PS, Bugge K, Marabini R, Boomsma W, Burow M, Kragelund BB
Protein Science, 2020. Vol.29, Iss. 1. doi:10.1002/pro.3754
Protein typically contain one or more domains with distinct structural properties and molecular functions that are retained when domains removed from the rest of the protein. In eukaryotic organisms, ca. 15 % of all proteins lack a defined tertiary structure and an even larger proportion comprises at least long (>50 residues) intrinsically disordered regions. The identification of disordered domains had been largely neglected so far. In an attempt to fill this gap, we used a sequence-based approach to assess and visualize domain organization in long intrinsically disordered regions based on compositional sequence biases.
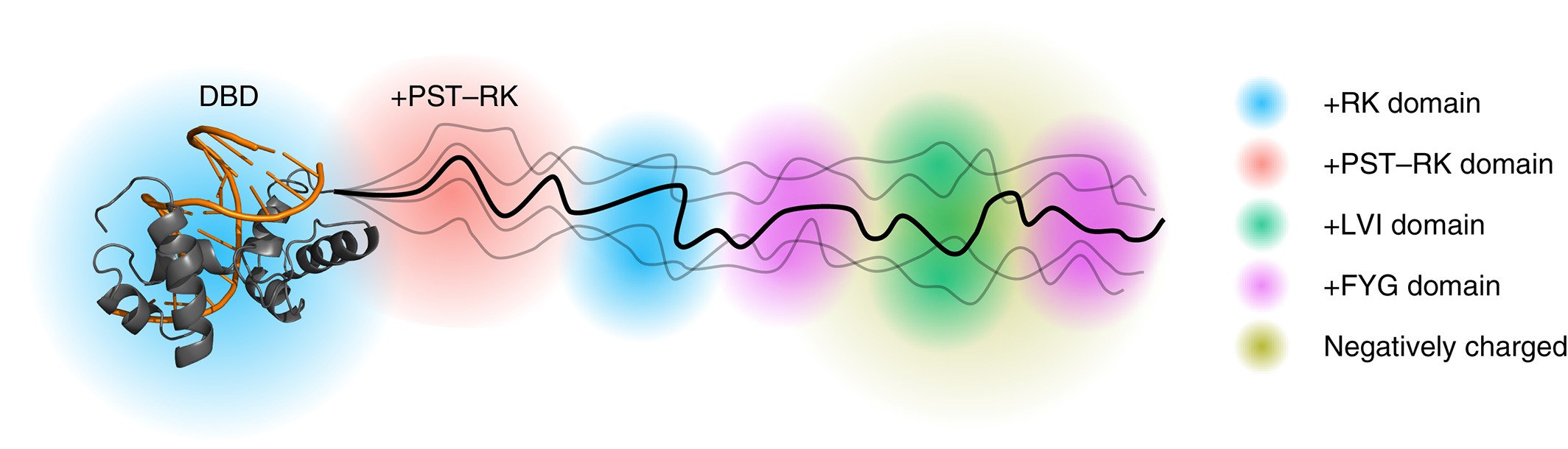 MYB DNA‐binding domain in complex with DNA with the protein coloured gray and DNA orange. Domains within the intrinsically disordered region have different sequence biases and thus distinct structural properties of relevance to differentiated molecular functions
MYB DNA‐binding domain in complex with DNA with the protein coloured gray and DNA orange. Domains within the intrinsically disordered region have different sequence biases and thus distinct structural properties of relevance to differentiated molecular functions
An online tool to find putative intrinsically disordered domains (IDDomainSpotter) in any protein sequence or sequence alignment is available at https://www.bio.ku.dk/sbinlab/IDDomainSpotter.
Using this tool, we have identified a putative domain enriched in hydrophilic and disorder-promoting residues (Pro, Ser, and Thr) and depleted in positive charges (Arg and Lys) bordering the folded DNA-binding domains of several transcription factors from plants and humans. Structural analyses of this domain showed the domain to be extended, dynamic and highly disordered. It connects the DNA-binding domain to other disordered domains and is present and conserved in several transcription factors from different families and domains of life.
This example illustrates the potential of IDDomainSpotter to predict, from sequence alone, putative domains of functional interest in otherwise uncharacterized disordered proteins.
